What is DNA?
DNA (Deoxyribonucleic Acid) is a long polymer of deoxyribonucleotides that stores genetic information in all living organisms. In most organisms DNA exists as a double-stranded helix formed by two complementary polynucleotide chains coiled around each other.
Examples of DNA content in organisms
- Bacteriophage Phi X 174: 5,386 nucleotides.
- Bacteriophage λ (lambda): 48,502 base pairs.
- Escherichia coli (E. coli): ≈ 4.6 × 10⁶ base pairs.
- Human haploid genome: ≈ 3.3 × 10⁹ base pairs.
Structure of the polynucleotide chain
Components of a nucleotide
A nucleotide consists of three components:
- A nitrogenous base (purine or pyrimidine)
- A pentose sugar (deoxyribose in DNA)
- A phosphate group
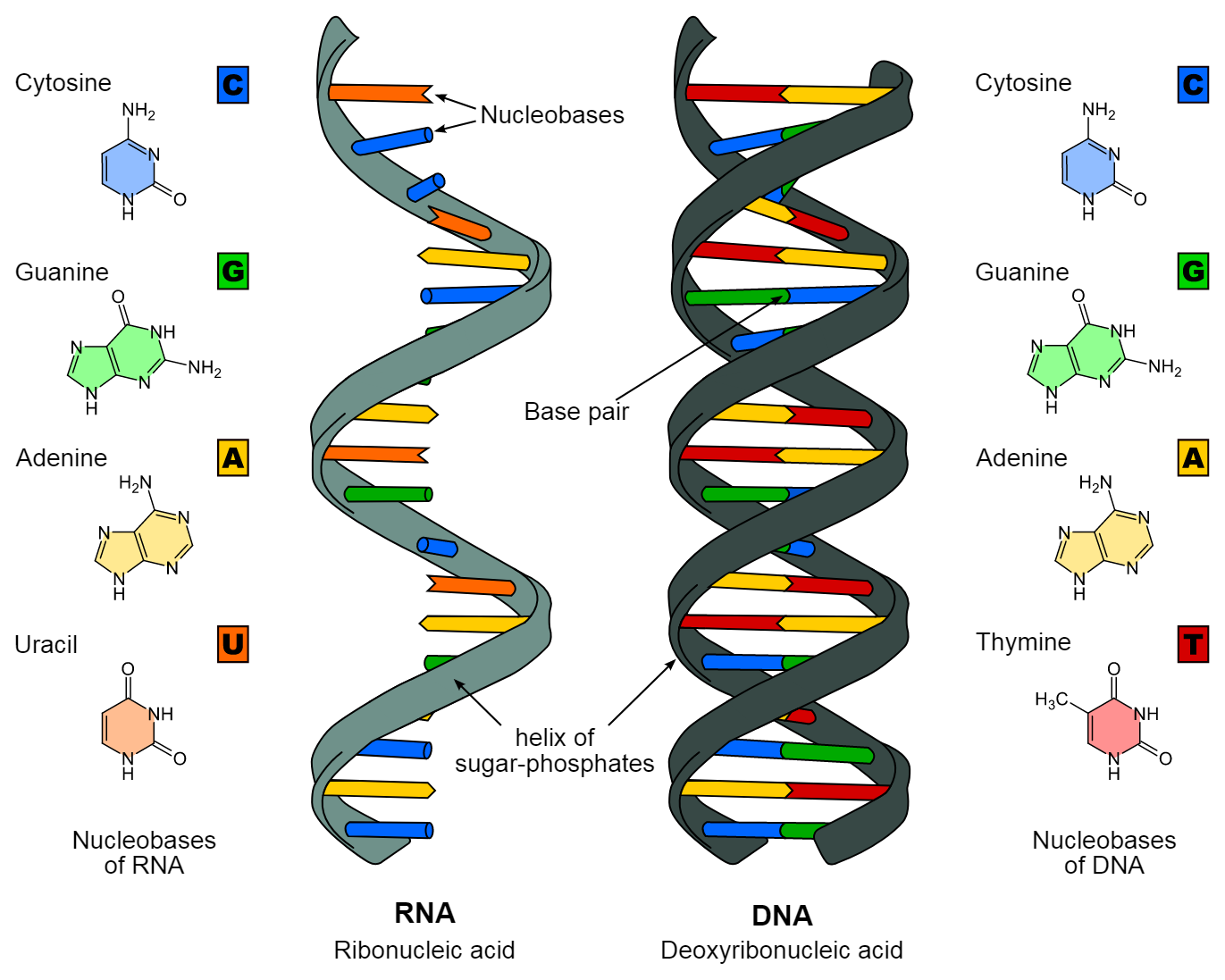
Types of nitrogenous bases
- Purines: Adenine (A) and Guanine (G)
- Pyrimidines: Cytosine (C) and Thymine (T) (Uracil (U) is found in RNA)
Cytosine occurs in both DNA and RNA; thymine is characteristic of DNA while uracil occurs in RNA.
Nucleosides and nucleotides
- A base + sugar = nucleoside (e.g. adenosine).
- Nucleoside + phosphate (attached at the 5′-OH) = nucleotide.
- Nucleotides join by 3′–5′ phosphodiester bonds to form a polynucleotide chain.
Each polynucleotide chain has directionality: a free phosphate at the 5′ end and a free hydroxyl at the 3′ end. The sugar–phosphate backbone gives structural support.
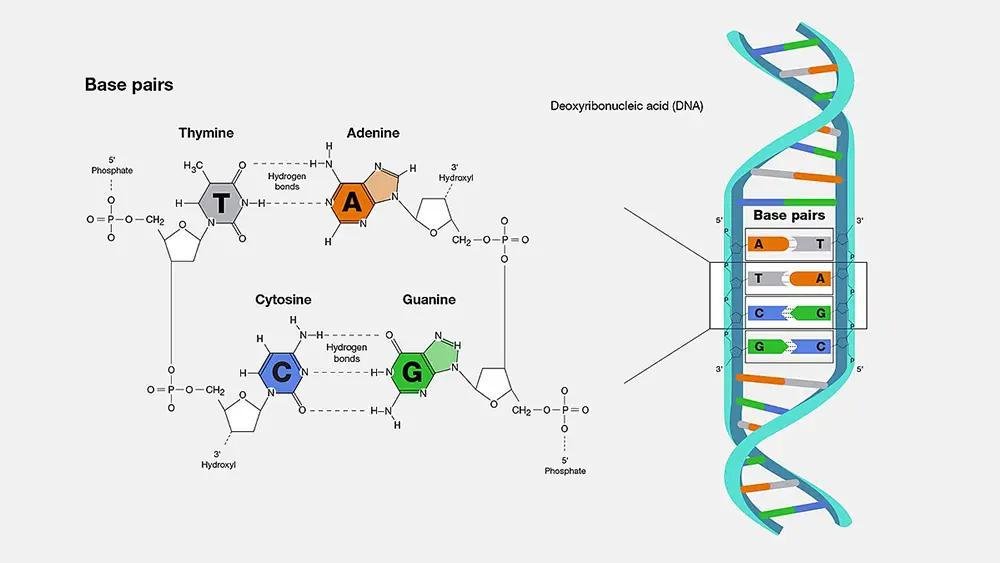
Base pairing and complementarity
Complementary base pairing is central to DNA function:
- A (adenine) pairs with T (thymine) via two hydrogen bonds.
- G (guanine) pairs with C (cytosine) via three hydrogen bonds.
Because of complementarity, the sequence of one strand determines the sequence of the other; this is vital for accurate replication.
Salient features of the double helix
- Two polynucleotide chains form the structure.
- The chains are antiparallel (one runs 5′→3′, the other 3′→5′).
- Base pairs are held by hydrogen bonds (A=T, C≡G).
- The helix is right-handed (B-DNA).
- One helical turn contains about 10 base pairs and the pitch (distance per turn) is ≈ 3.4 nm.
- Distance between adjacent base pairs ≈ 0.34 nm.

Packaging of the DNA helix
Although the DNA double helix in a typical mammalian cell is about 2.2 metres long, it fits into a nucleus only a few micrometres across because of hierarchical packaging.
In prokaryotes (e.g. E. coli)
- DNA is not membrane-bound; it is organised in a nucleoid with looped domains, aided by DNA-binding proteins and supercoiling.
In eukaryotes (e.g. humans)
- DNA wraps around histone proteins forming nucleosomes (“beads on a string”).
- Nucleosomes fold into higher-order structures to form chromatin, which condenses into chromosomes during cell division.
This organisation allows DNA to be compact yet accessible for replication and transcription.
Importance of DNA
- Genetics & inheritance: DNA stores hereditary information and directs protein synthesis that determines traits.
- Medicine: DNA analysis enables genetic testing, diagnosis, forensics (DNA fingerprinting), and gene therapy research.
- Biotechnology: Recombinant DNA technology produces medicines (e.g. insulin), vaccines and GM crops.
- Research & society: Projects like the Human Genome Project have advanced personalized medicine and our understanding of biology.
Exercises
Exercise A — Short problems (SET A adapted)
- DNA duplication in coli takes 20 minutes. An E. coli cell with heavy DNA is allowed to grow in ¹⁴N-containing NH₄Cl for 100 minutes. Find the ratio of hybrid DNA molecules to light molecules after 100 minutes. Draw the result (explain reasoning).
- In a linear dsDNA molecule of 100 base pairs, 30% is guanine. Find the total number of hydrogen bonds and phosphodiester bonds in that DNA.
- If the antisense strand of DNA (3′→5′) has the following sequence: 3′–T A C C C G A C G A T C–5′, find the sequence of its cDNA and of the mRNA.
- coli has 4.6 × 10⁶ bp in its DNA. Find the length (in metres) of E. coli DNA.
- Human haploid genome has 3.3 × 10⁹ bp. Find the length (in metres) of human haploid DNA.
- If the pitch of B-DNA is 3.4 nm per 10 bp, how many base pairs are found within 34 nm?
- Given an mRNA strand 5′–AUG CCC AAA UUU UUA UAA–3′, find how many amino acids are formed from this sequence; which end of tRNA carries the amino acid, and how many tRNA molecules are required?
- (Variation of Q1) Same as Q1 but growth period = 80 minutes. Compute the ratio of hybrid to light DNA molecules after 80 minutes.
- In a linear dsDNA molecule of 100 base pairs, 20% is guanine. Find the total number of hydrogen bonds and phosphodiester bonds.
- If the antisense strand (3′→5′) is 3′–T A C C C G A C G A T C C C A A A–5′, find the cDNA and mRNA sequences.
Answers & worked solutions
General notes for answers: E. coli DNA replication is semi-conservative. Each replication cycle (20 minutes) doubles the number of DNA molecules; after nnn cycles there are 2n2^n2n total DNA molecules. “Heavy” vs “light” refers to parental (heavy) vs newly synthesised (light) strands; hybrid molecules contain one heavy and one light strand.
- After 100 min:
Replication time = 20 min → cycles = 100/20=5100/20 = 5100/20=5 cycles. Total molecules = 25=322^5 = 3225=32. Semi-conservative replication: after 1 cycle all molecules are hybrid (1 heavy + 1 light strand). After 2 cycles some become light–light, and so on. The number of hybrid molecules after nnn cycles = 2n−12^{n-1}2n−1. So for n=5n=5n=5, hybrid = 24=162^{4} = 1624=16. Light (completely light–light) molecules = total − hybrid − parental (if parental remains) but parental heavy–heavy are 0 after first round (parental distributes). Standard semi-conservative result: after nnn cycles hybrid = 2n−12^{n-1}2n−1, light = 2n−1−2^{n-1}-2n−1− (but simpler: after 5 cycles hybrid = 16, light = 16). So ratio hybrid : light = 1 : 1 (16:16).
(Graphic: 32 circles — 16 hybrid, 16 light.) - 100 bp, 30% G:
G = 30 bp → C = 30 bp (complementary). Remaining bases = 40 bp → A = 20 bp, T = 20 bp. Hydrogen bonds: each A–T = 2 bonds; each G–C = 3 bonds. Total H bonds = (20×2)+(30×3)=40+90=130 (20\times2) + (30\times3) = 40 + 90 = 130(20×2)+(30×3)=40+90=130 hydrogen bonds (per double helix). Phosphodiester bonds: each strand of 100 bases has 99 phosphodiester bonds; two strands → 2×99=1982 \times 99 = 1982×99=198 phosphodiester bonds. - Antisense (given 3′→5′): 3′–T A C C C G A C G A T C–5′
First write antisense 5′→3′: 5′–C T A G C A G C C C A T–3′ (reverse the order). But easier method: cDNA (complement of antisense) will be the sense/coding strand (5′→3′). The coding strand (cDNA) sequence is: 5′–A T G G G C T G C T A G–3′ (which corresponds to original). mRNA (transcribed from coding strand replacing T with U): 5′–A U G G G C U G C U A G–3′. (Translate accordingly: starts at AUG). - Length of E. coli DNA:
Number of bp = 4.6 × 10⁶. Each bp rise ≈ 0.34 nm = 0.34 × 10⁻9 m. Length = 4.6×106×0.34×10−94.6\times10^6 \times 0.34\times10^{-9}4.6×106×0.34×10−9 m = 1.564×10−31.564\times10^{-3}1.564×10−3 m = 1.564 mm. - Length of human haploid DNA:
3.3 × 10⁹ bp × 0.34 × 10⁻9 m = 1.122×1001.122\times10^01.122×100 m ≈ 1.12 metres (per haploid set).
**6. Pitch 3.4 nm per 10 bp ⇒ 34 nm contains (34 / 3.4) × 10 bp = 10 × 10 = 100 bp.
- mRNA 5′–AUG CCC AAA UUU UUA UAA–3′
Translate into codons: AUG | CCC | AAA | UUU | UUA | UAA
- AUG = Met (start, also codes Met).
- CCC = Pro
- AAA = Lys
- UUU = Phe
- UUA = Leu
- UAA = Stop (termination)
So amino acids formed before stop: Met, Pro, Lys, Phe, Leu → 5 amino acids.
tRNA carries amino acid on its 3′ end (the CCA acceptor end). One tRNA molecule is required for each amino acid added (so up to 5 tRNAs will be used during translation of these five codons, though the initiating Met tRNA is reused in other translations).
- After 80 min: cycles = 80/20 = 4 → total molecules =24=16=2^4=16=24=16. Hybrid molecules =23=8=2^{3}=8=23=8. Light molecules =8=8=8. Ratio hybrid:light = 1:1.
- 100 bp, 20% G → G = 20, C = 20, remaining 60 → A = 30, T = 30.
Hydrogen bonds = (30×2)+(20×3)=60+60=120 (30\times2) + (20\times3) = 60 + 60 = 120(30×2)+(20×3)=60+60=120. Phosphodiester bonds = 2×99=1982\times99 = 1982×99=198. - Antisense 3′–T A C C C G A C G A T C C C A A A–5′
cDNA (coding strand, 5′→3′) = 5′–A T G G G C T G C T A G G G T T T–3′
mRNA = 5′–A U G G G C U G C U A G G G U U U–3′
Short revision checklist (quick facts)
- Base pair distance = 0.34 nm.
- 10 bp ≈ 3.4 nm (one helical turn).
- A–T = 2 H bonds, G–C = 3 H bonds.
- Direction: strands are antiparallel (5′ ↔ 3′).
- Human haploid DNA length ≈ 1.12 m.